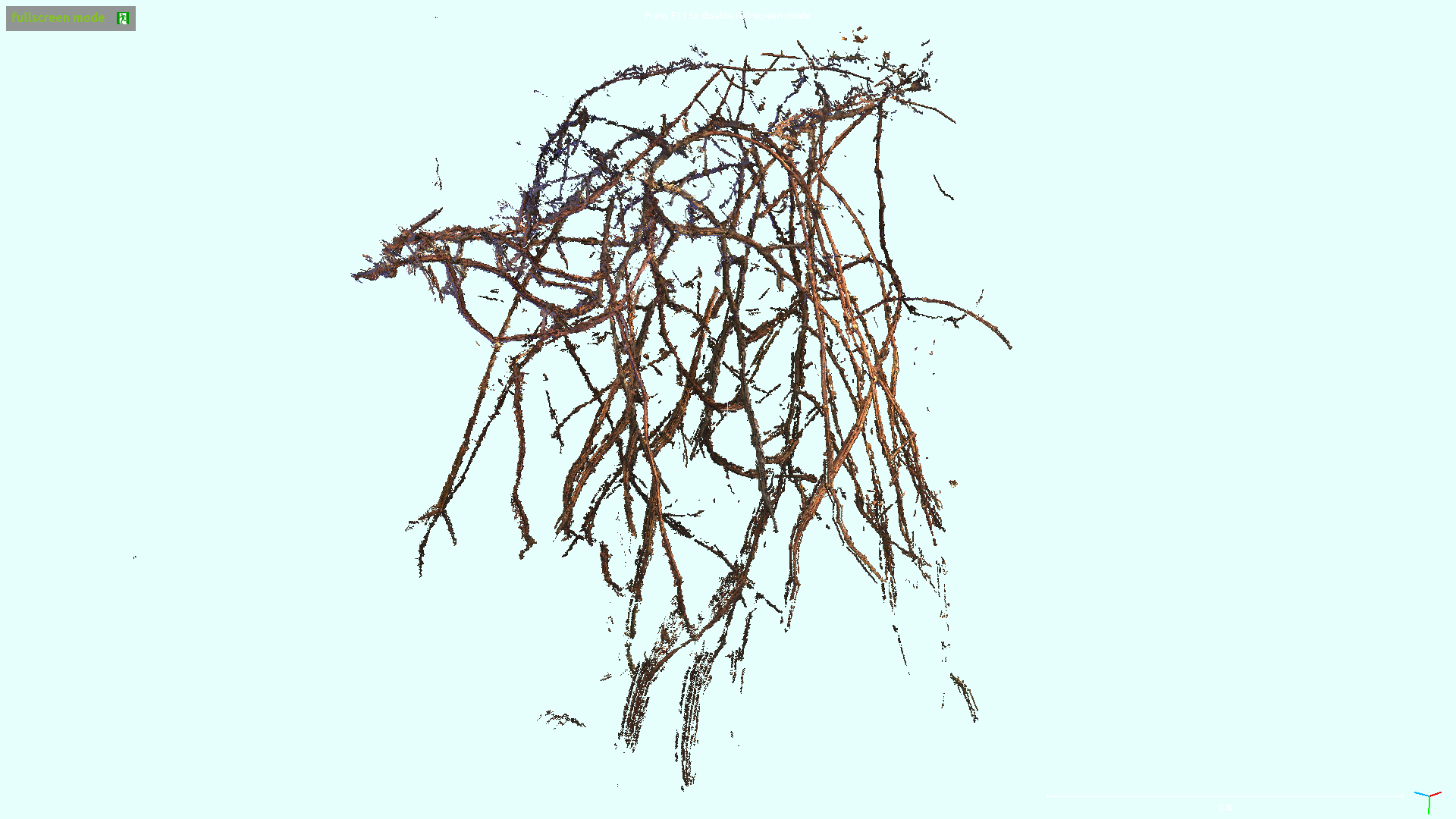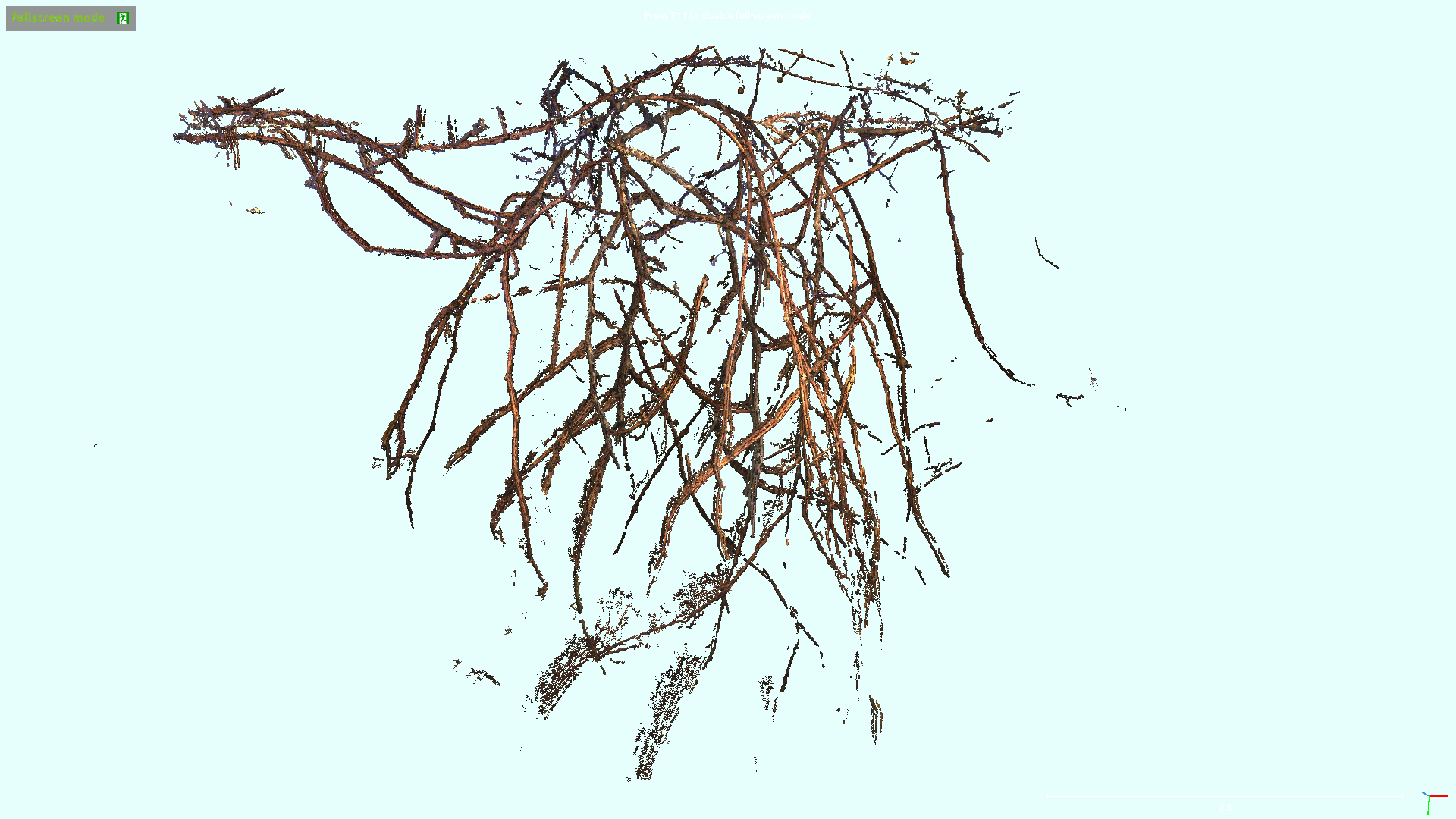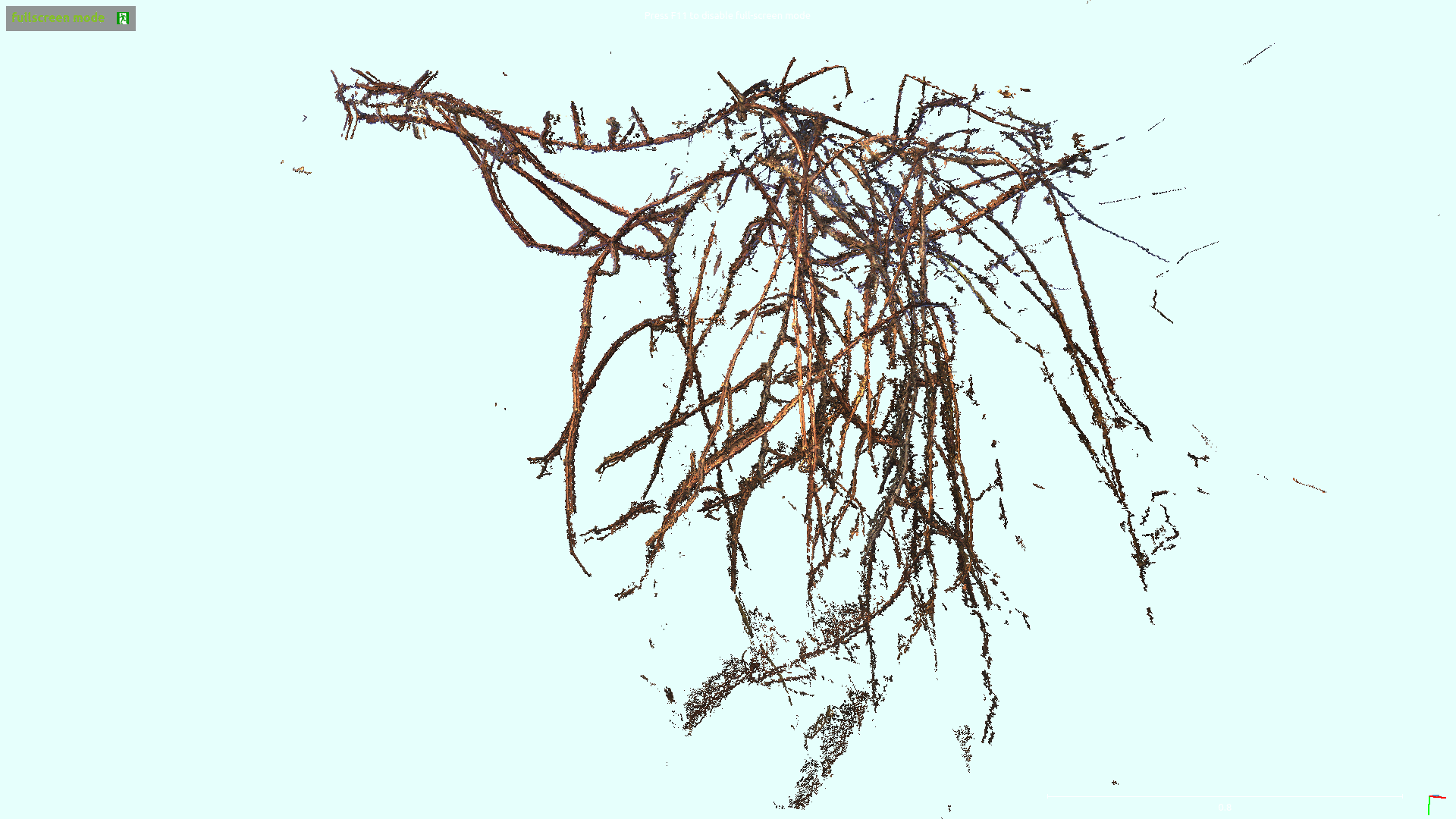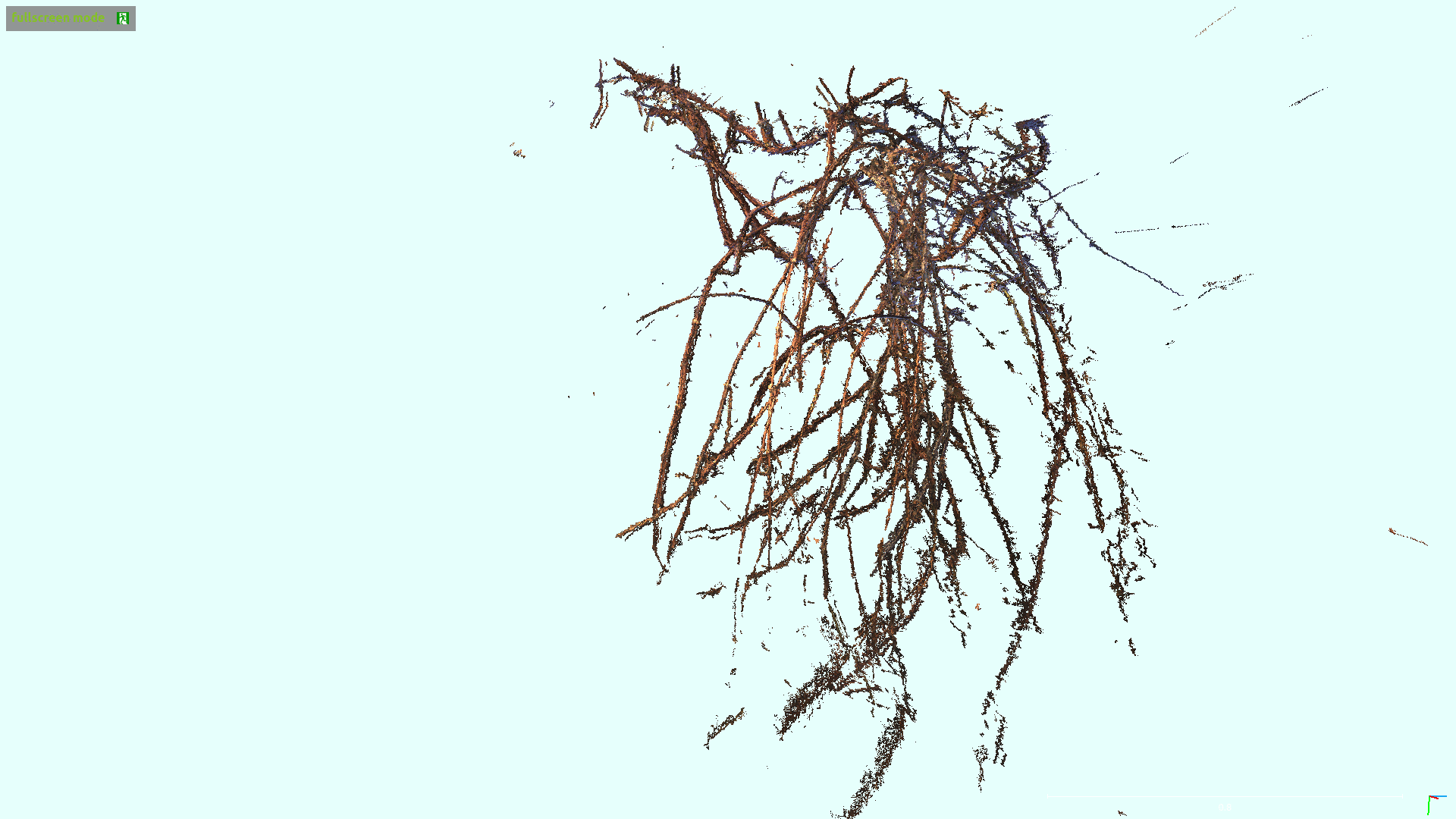3D Skeletonization of Complex Grapevines
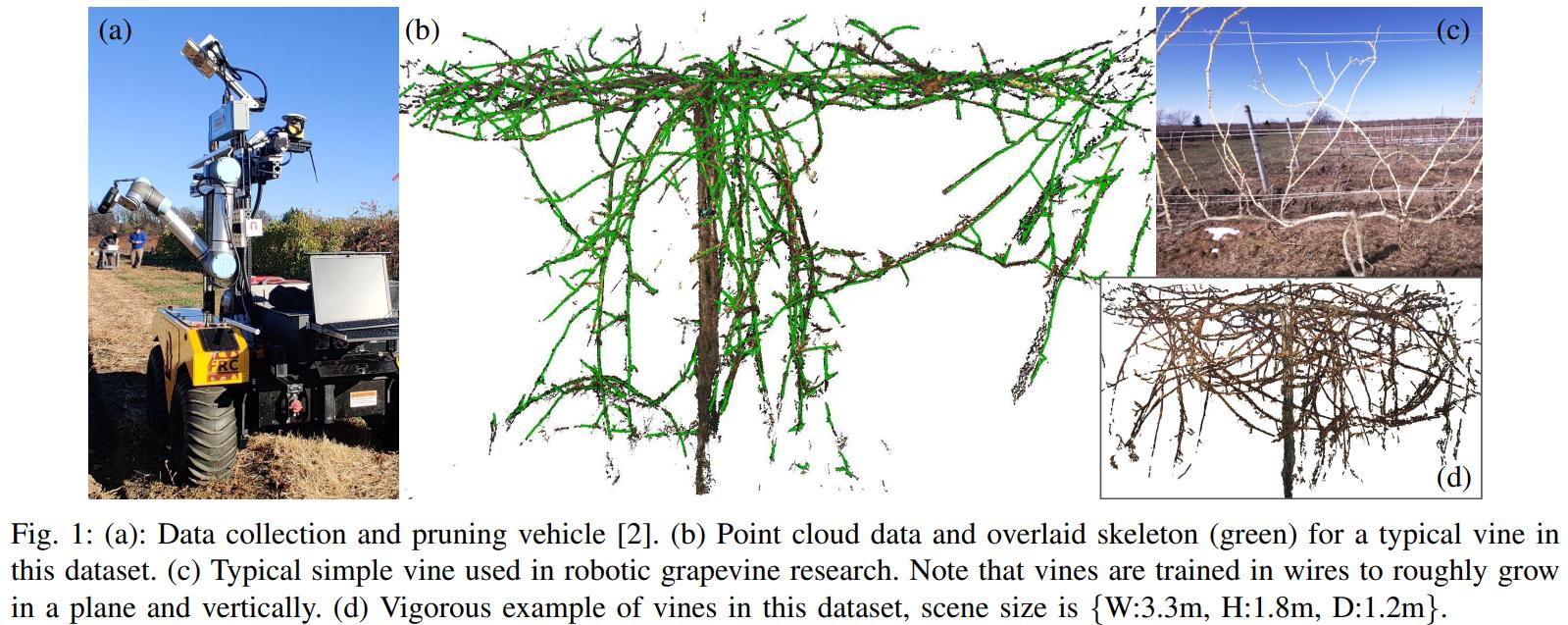
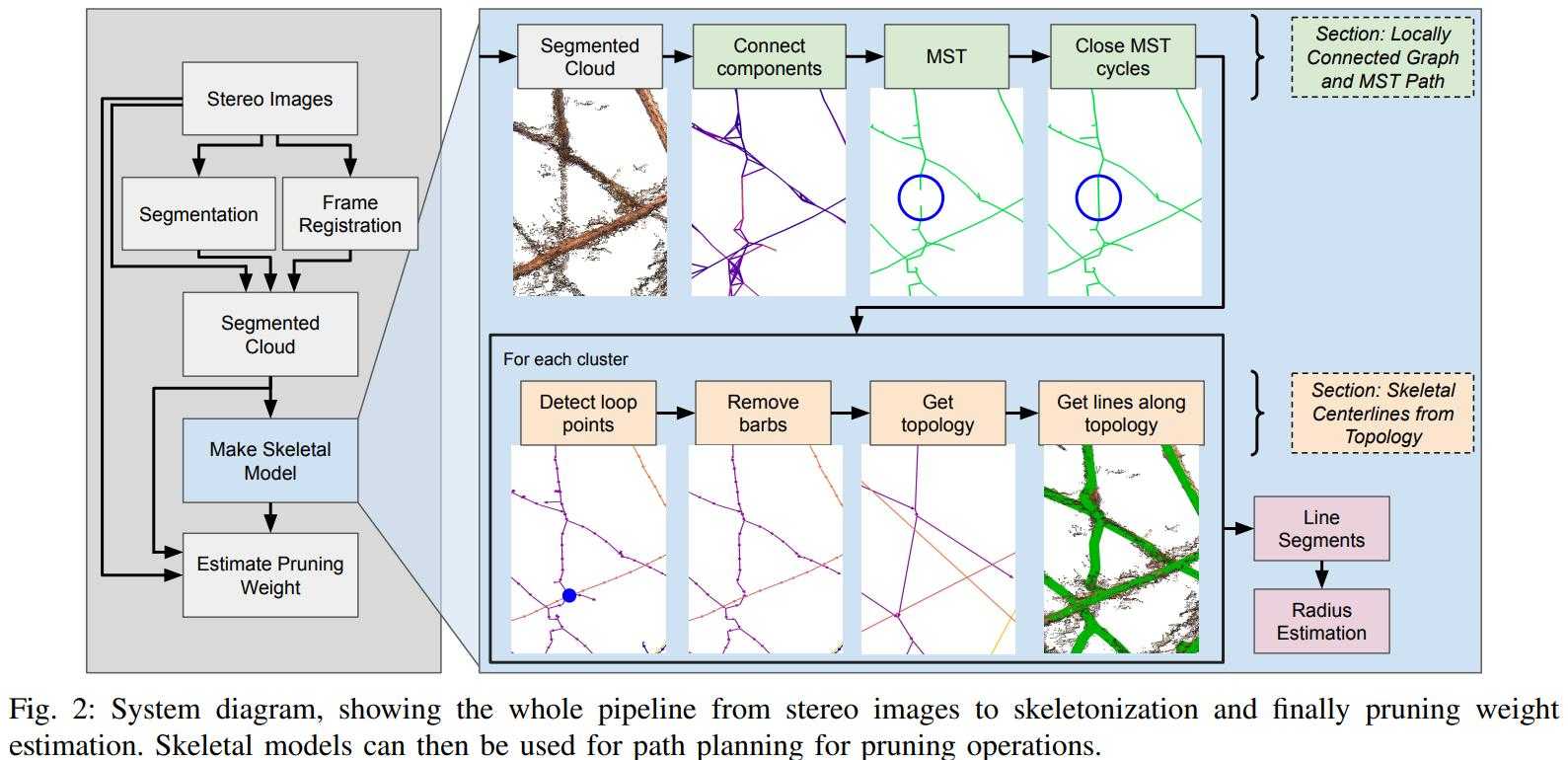
In the Kantor Lab at CMU, surveying fields to sample crop phenotypes for breeding or farm management was a task of interest. In this particular project, we tested the efficacy of reconstructing the grapevines from vehicular side-mounted stereo images, and then did our best to estimate skeletal structure and pruning weight from the reconstructions. Generally speaking I adjusted the 3D points into alignment via ICP, ran a 2D vine segmentation model to identify vine points, and then went through a graph refinement process to estimate the skeletal size and position. For more information you can check out the paper on IEEE or arXiv. This paper was accepted at IROS 2023.
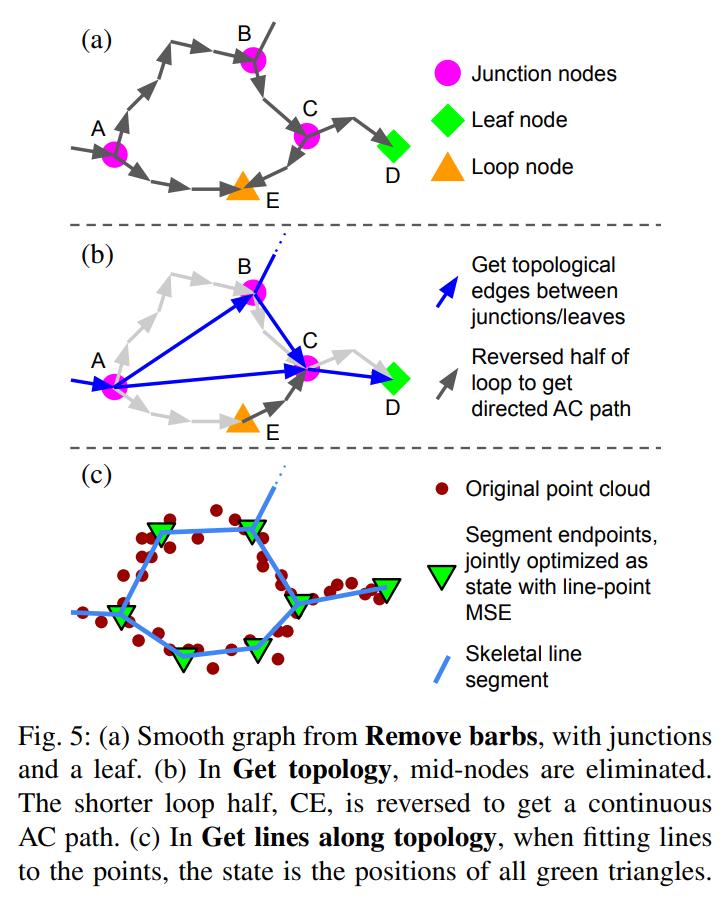
Overall I would say the final process was a bit too long to make me really satisfied. Whenever you have a complex set of steps it invites fagility, because you have a series of failure points where a new dataset could violate assumptions made at any of the stages. Perhaps with development this could be shorted and simplified. I was happy with the method for estimating skeletal radii. By taking a particular matrix construction I was able to estimate the the radii of all vines in a simple manner, where prior radius assumptions, a smoothing term, and a data-fitting term were all included. Although this must be inspired from other sources I don’t remember drawing from a particular place, I believe I constructed it myself, and the cleanliness of the approach was satisfying.
As preparation I looked into a variety of papers about methods for getting tree skeletons out of point cloud data. See presentation for a literature review.